Before container based software has been developemented, HPC or High Performance Computing system, administrators use to compile application manually solving libary dependency issues and config applications with environment module. User can load part of softwares as they are only needed. The advantages of environment modules are that they allow you to load and unload software configurations dynamically in a clean fashion, providing end users with the best experience when it comes to customizing a specific configuration for each application.
1
2
3
$ module avail
$ module load gromacs
$ gmx ...
However, wildly satifying user needs for distributed system, HPC and AI applications, complex library dependenies is nighmare for administor. HPC administrators are often overwhelmed with the amount of time they must spend to install, upgrade, and monitor software. HPC system is slowly upgraded system in user point view for HPC. So organization to Public HPC public cloud.
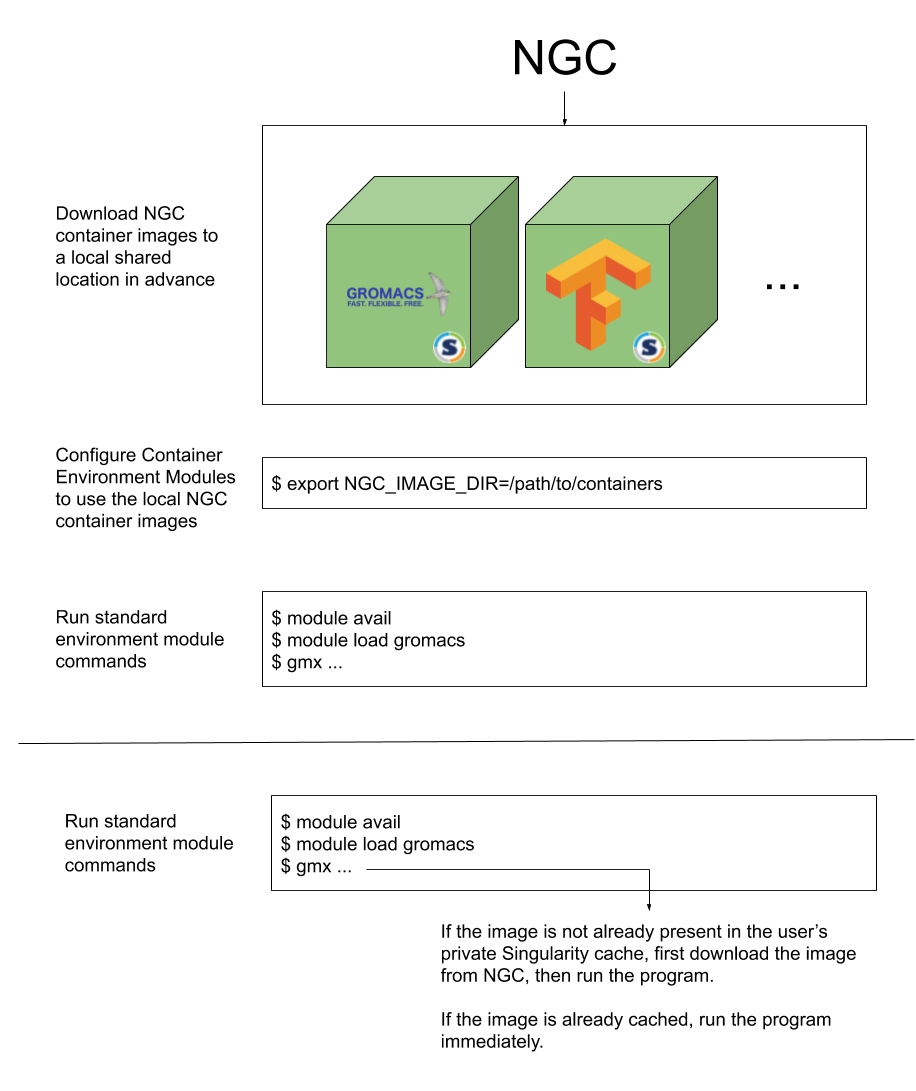
Docker or Singularity container can be a great way to simplify the overall development and deployment process, or DevOps. DockerHub, public share dockerfile and NVIDIA Container NGC provide hurge pre-compile and pre-configure for HPC user. If some journal paper provide paper with code which is usaully come with container to repeat the resarch results. In this post, we motivate you to apply NVIDIA NGC Container with Environment Modules, which jointed together to make our process faster and more easier.
Why containers?
- Allow you to
package a softwareapplication, libraries, and other runtime dependencies into a single image Eliminate the need to install complex software environments- Enable you to pull and
run applications on the system without any assistance from system administrators - Allow researchers to
share their applicationwith other researchers for corroboration
Source: https://developer.nvidia.com/blog/simplifying-hpc-workflows-with-ngc-container-environment-modules/
Container as Modules
The past few years, we have seen containers become quite popular when it comes to deploying HPC applications,such as BioContainers. However, most super computing center and users submit job or running application using environment modules.
Use familiar environment module commands, ensuring a minimal learning curve or change to existing workflows
Make use of all the benefits of containers, including and reproducibility
Leverage all of the optimized HPC and Deep Learning containers from Docker Hub, NVIDIA Container
Following step is initial process that setup only time first time.
Outline
Guide from ngc-container-environment
1
2
3
4
5
6
7
8
$ ssh username@aim1.mahidol.ac.th
$ salloc -w some_compute_node
$ ssh username@some_compute_node
$ git clone https://github.com/NVIDIA/ngc-container-environment-modules
$ module use $(pwd)/ngc-container-environment-modules
$ module load gromacs
$ gmx
The last two steps are runing jobs. The gmx command on the host is linked to the GROMACS container. Singularity mounts $HOME,/tmp and $(PWD) by default.
Custmize Configure for Exascale Cluster
The NGC container environment modules are a reference. It is expected that they will need some modification for Exascale cluster.
The name of the Singularity module. The container environment modules try to load the
Singularitymodule (note theb capital ‘S’). Set theNGC_SINGULARITY_MODULEenvironment variable set it tonone.Singularity uses a temporary directory to build the squashfs filesystem, and this temp space needs to be large enough to hold the entire resulting Singularity image. By default this happens in
/tmpbut the location can be configured by settingSINGULARITY_TMPDIR
1
2
3
4
5
6
7
8
9
10
$ echo '# Setup Singularity and Module work together' >> ~/.bashrc
$ echo 'export SINGULARITY_TMPDIR="/home/snit.san/tmp"' >> ~/.bashrc
$ echo 'export NGC_SINGULARITY_MODULE=none' >> ~/.bashrc
$ echo 'module use /home/snit.san/ngc-container-environment-modules' >> ~/.bashrc
$ echo 'export SINGULARITY_TMPDIR=/home/snit.san/tmp' >> ~/.bashrc
Examples
Basic : One Node Multi GPUs
Download a GROMACS benchmark to run this exmaple.
1
2
3
4
5
6
7
8
9
10
$ wget https://ftp.gromacs.org/pub/benchmarks/water_GMX50_bare.tar.gz
$ tar xvfz water_GMX50_bare.tar.gz
$ salloc -w turing
$ ssh turing
$ cd /home/snit.san/GROMACS/water-cut1.0_GMX50_bare/1536
$ module load gromacs/2020.2
$ gmx mdrun -ntmpi 1 -ntomp 40 -v -pin on -nb gpu --pme gpu --resetstep 12000 -nsteps 20000 -nstlist 400 -noconfout -s topol.tpr
Interactive : Deep Learningg with PyTorch
1
2
3
4
5
6
7
8
9
10
11
12
13
$ salloc -w zeta
$ ssh zeta
$ module load pytorch/20.06-py3
$ python3
>>> import torch
>>> x = torch.randn(3, 7)
>>> x
tensor([[-0.5307, 0.8221, -0.8459, 0.7091, -0.7165, 0.6743, 1.8597],
[ 0.0961, 0.3864, 0.6767, -1.4271, 1.4069, -0.4359, -2.3566],
[-0.4707, -1.0400, 0.2112, -0.3847, -1.6868, 0.4977, -0.1213]])
>>>
Jupyter notebooks: cuSignal possible with Rapids project
1
2
3
4
$ salloc -w turing
$ ssh turing
$ module load rapidsai
$ jupyter notebook --ip 0.0.0.0 --no-browser --notebook-dir /rapids/notebooks
Multi Node Multi GPUs: MNMG with Kokkos and MPI
Download the LAMMPS Molecular Dynamics Simulator, Lennard Jones fluid dataset to run this example.
1
2
3
4
5
6
$ mkdir LAMMPS_test
$ cd LAMMPS_test
$ wget https://www.lammps.org/inputs/in.lj.txt
$ module load lammps/15Jun2020
$ mpirun -n 2 lmp -in in.lj.txt -var x 8 -var y 8 -var z 8 -k on g 2 -sf kk -pk kokkos cuda/aware on neigh full comm device binsize 2.8